How to automate R reports on Linux with cron jobs
You can find the code that we work through in this example here.
The motivation might be something like this: You have some data that is updated at regular intervals (say, daily). The data is held in some database, or perhaps it's an Excel file on SharePoint, or someone manually updates a Google Sheet every day. You've written some data processing pipeline in R that pulls the data; transforms it; creates some sort of report (maybe a PDF or HTML file in R Markdown, or perhaps you're just outputting cleaned data files); and then uploads the report to some server, or attaches it in an email to the client.
Since you're a programmer (or an analyst or a data scientist or whatever you want to call yourself), you're perfectly happy spending hours on end tinkering with something in order to save yourself a few minutes of time down the track. So, you want to automate the whole thing. That's where cron jobs on Linux can help.
This whole process is fairly straightforward, but there are definitely a few details that can catch you out. Before we start, it would be silly not to mention the very useful R packages cronR (for Unix) and taskscheduleR (for Windows). Instead of using these packages, we're going to be creating cron jobs manually. Either method is fine; I just like the idea of becoming more acquainted with cron, because that'll make automating any other task on Linux much easier. And we can pretend to be sysadmins.
Setting up
If you're reading this blog post you're probably on a Linux machine, so open the terminal and paste in the following:
mkdir ~/cron-reporting-example
cd ~/cron-reporting-example
mkdir data && mkdir docs && mkdir output && mkdir scripts
echo "write.csv(iris, 'output/new_iris.csv')" > scripts/new_iris.R
touch master.R
rstudio
This creates a project directory cron-reporting-example
in your home path; a bunch of project-related folders that I tend to
use for everything; a minimal example script new_iris.R that writes the iris
dataset into an output folder; and a master script that will source the iris-writing script.
We then open RStudio. I'm assuming
you have RStudio installed and it's run by rstudio; if
you're on a server then do what you gotta do. Finally,
create a new R project file for that directory.
Basic logging
Let's open up the R script master.R. The first thing
we're going to add is some logging! It's easy to forget this basic step,
but you'll be glad you set it up, trust me.
We're going to make our cron job gather up any messages printed
to the R console and output them to a log file. So, anything we print
will be captured. I prefer to use cat rather than
print for this, as the latter will wrap everything in quotes.
We can start with something like this:
cat(paste0(Sys.time(), " Starting cron job...\n"))Pretty simple: print the date and time, along with a brief message. We can finish with an optional newline. I like to add messages like this before every new piece of the puzzle. For example, I might have a message that says we're about to source the script that downloads data; a message that says we're about to source the R Markdown script that produces the report; and so on. Doesn't have to be too technical, just little milestones in the ground so that if something goes wrong, you have a better idea of where that might have happened.
Sourcing scripts
We can source a script in the usual way. Add this to your master.R script:
cat(paste0(Sys.time(), " Sourcing scripts/new_iris.R...\n"))
source("scripts/new_iris.R")
cat(paste0(Sys.time(), " Finished running scripts/new_iris.R.\n"))
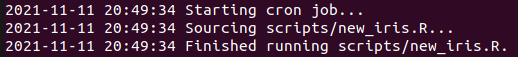
Rscript master.R in the terminal, we'll see
the logging (and the R code) printed, and there should be a new_iris.csv
file in the output folder:
if (!interactive()) setwd("~/cron-reporting-example")This goes at the top of master.R.
Basic crontab
This post assumes that you know what a crontab is; I highly recommend this post if you're interested in a comprehensive introduction. And for help with cron schedule expressions, crontab guru is amazing.
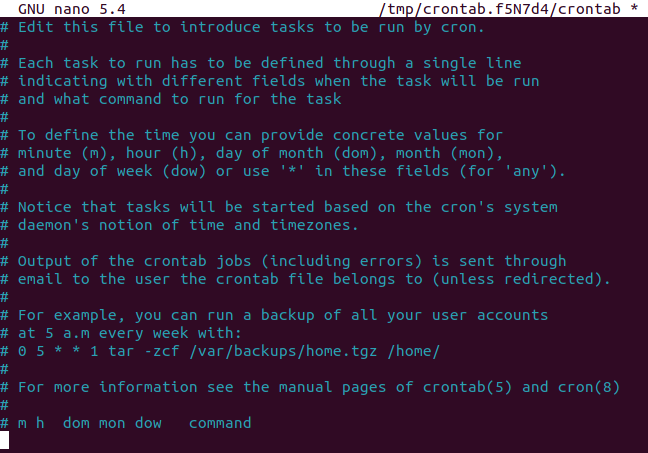
So let's edit our crontab, by running crontab -e
in the terminal. If you're a peasant like me and you use nano,
it should look something like this:
There are a few key things we need to include in our cron expression. Let's go through them one by one.
- Schedule expression: How often should the job be run?
For testing, I usually create a job that runs every minute. (As
long as the script itself runs in a few seconds, this will be fine!)
That's the easiest schedule expression: just
* * * * *. - Command to run: This will be the
Rscriptbinary. You can provide a specific binary if you have a particular version of R in mind; I'm just usingRscript. - Path to script to run: This is the full path to the R script
that we actually want to run. For us, this will be
~/cron-reporting-example/master.R. - Output to log file: This one is optional, but it's
definitely a good idea to include it. We can create a .log file that
is analogous to the
master.Rscript at the same level, and pass it in like this:>> ~/cron-reporting-example/master.log 2>&1.>>is the Bash append operator.x >> yappends the output ofxinto the filey.2>&1is a definition of what type of output to pass to the log file. I'm just using what I've seen elsewhere—haven't given it too much thought, but that seems to do the trick.
So this is what our final crontab will look like:
* * * * * Rscript ~/cron-reporting-example/master.R >> ~/cron-reporting-example/master.log 2>&1
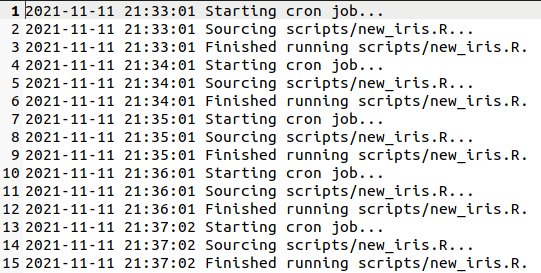
Paste that into your crontab, save and close it, and... wait one minute. Take
a look at the modified time of output/new_iris.csv—if everything
is working, it should
update every minute! You should also have a log file at ~/cron-reporting-example/master.log,
which will also update every minute and look something like this:
Okay! At this point I'd recommend deleting the crontab, so that you aren't continually calling a pointless R script (and appending useless information into a logfile) every minute...
Next steps
The next thing you're probably wondering is how to wire this up with an R Markdown script. That's also pretty straightforward, but there are some surprises in how the filepaths work when you're rending a .Rmd file from an R script... But we'll leave that for a future update.